Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem
Automated segmentation of anatomical structures is a crucial step in image analysis. For lung segmentation in computed tomography, a variety of approaches exist, involving sophisticated pipelines trained and validated on different datasets. However, the clinical applicability of these approaches across diseases remains limited. We compared four generic deep learning approaches trained on various datasets and two readily available lung segmentation algorithms. We performed evaluation on routine imaging data with more than six different disease patterns and three published data sets. Using different deep learning approaches, mean Dice similarity coefficients (DSCs) on test datasets varied not over 0.02. When trained on a diverse routine dataset (n = 36) a standard approach (U-net) yields a higher DSC (0.97 $\pm$ 0.05) compared to training on public datasets such as Lung Tissue Research Consortium (0.94 $\pm$ 0.13, p = 0.024) or Anatomy 3 (0.92 $\pm$ 0.15, p = 0.001). Trained on routine data (n = 231) covering multiple diseases, U-net compared to reference methods yields a DSC of 0.98 $\pm$ 0.03 versus 0.94 $\pm$ 0.12 (p = 0.024).
PDF Abstract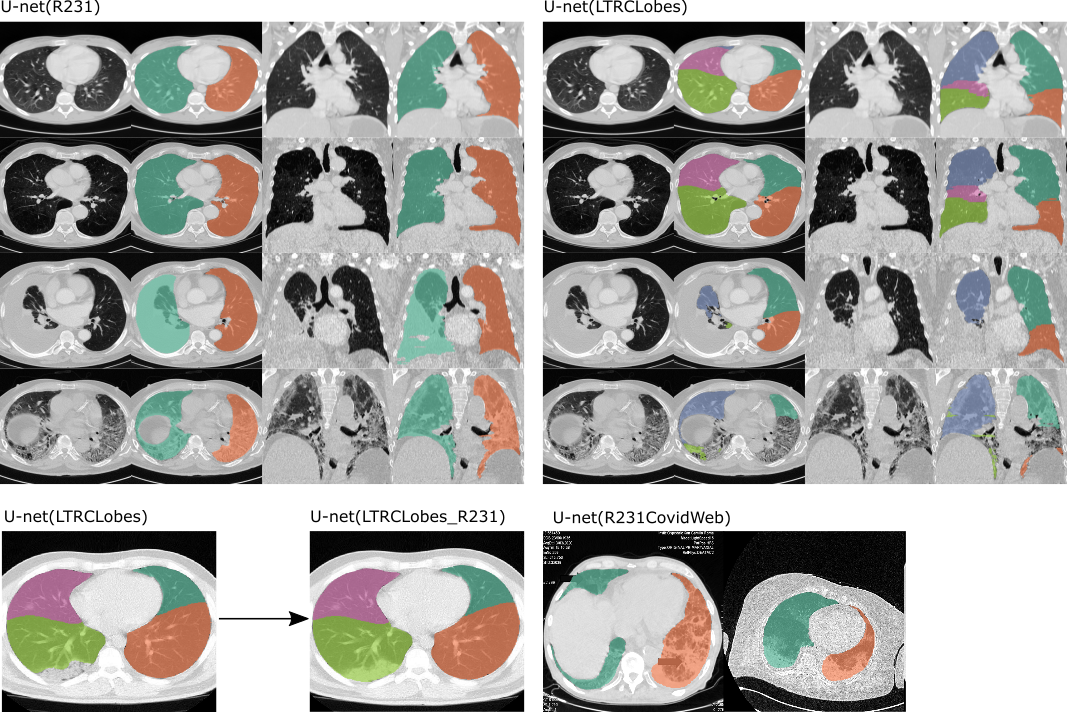



 100DOH
100DOH