KiU-Net: Towards Accurate Segmentation of Biomedical Images using Over-complete Representations
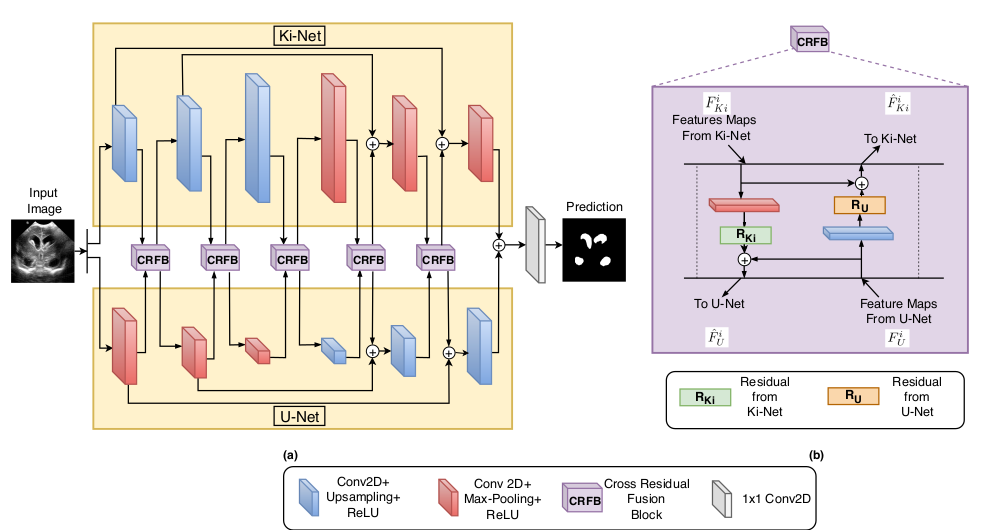
Due to its excellent performance, U-Net is the most widely used backbone architecture for biomedical image segmentation in the recent years. However, in our studies, we observe that there is a considerable performance drop in the case of detecting smaller anatomical landmarks with blurred noisy boundaries. We analyze this issue in detail, and address it by proposing an over-complete architecture (Ki-Net) which involves projecting the data onto higher dimensions (in the spatial sense). This network, when augmented with U-Net, results in significant improvements in the case of segmenting small anatomical landmarks and blurred noisy boundaries while obtaining better overall performance. Furthermore, the proposed network has additional benefits like faster convergence and fewer number of parameters. We evaluate the proposed method on the task of brain anatomy segmentation from 2D Ultrasound (US) of preterm neonates, and achieve an improvement of around 4% in terms of the DICE accuracy and Jaccard index as compared to the standard-U-Net, while outperforming the recent best methods by 2%. Code: https://github.com/jeya-maria-jose/KiU-Net-pytorch .
PDF Abstract